Run a random sub-sampling modification of the SDAR algorithm as
standardized in "ASTM E3076-18". As the original version uses numerous
linear regressions (.lm.fit() from the stats-package), it can be
painfully slow for test data with high resolution. The lazy variant of the
algorithm will use several random sub-samples of the data to find an
estimate for the fit-range within the data and thus can improve processing
speed. See the article Speed Benchmarking the SDAR-algorithm
for further information.
Usage
sdar_lazy(
data,
x,
y,
verbose = TRUE,
plot = TRUE,
n.fit = 5,
cutoff_probability = 0.5,
...
)Arguments
- data
Data record to analyze. Labels of the data columns will be used as units.
- x, y
<
tidy-select> Columns with x and y within data.- verbose, plot
Give a summarizing report / show a plot of the final fit.
- n.fit
Repetitions of drawing a random sub-sample from the data in the normalized and finding a fitting range to find an estimate for the final fitting range.
- cutoff_probability
Cut-off probability for estimating optimum size of sub-sampled data range via logistic regression, which is predicting if sub-sampled data will pass the quality checks.
- ...
<
dynamic-dots> Pass parameters to downstream functions: e.g. setverbose.allorplot.alltoTRUEto get additional diagnostic information during processing data. Setenforce_subsamplingtoTRUEto run the random sub-sampling algorithm even though it might be slower than the standard SDAR-algorithm.
Value
A list containing a data.frame with the results of the final fit, lists with the quality- and fit-metrics, and a list containing the crated plot-functions.
Note
The function can use parallel processing via the
furrr-package. To use this feature, set
up a plan other than the default sequential strategy beforehand. Also, as
random values are drawn, set a random seed beforehand to get
reproducible results.
References
Lucon, E. (2019). Use and validation of the slope determination by the analysis of residuals (SDAR) algorithm (NIST TN 2050; p. NIST TN 2050). National Institute of Standards and Technology. https://doi.org/10.6028/NIST.TN.2050
Standard Practice for Determination of the Slope in the Linear Region of a Test Record (ASTM E3076-18). (2018). https://doi.org/10.1520/E3076-18
Graham, S., & Adler, M. (2011). Determining the Slope and Quality of Fit for the Linear Part of a Test Record. Journal of Testing and Evaluation - J TEST EVAL, 39. https://doi.org/10.1520/JTE103038
See also
sdar() for the standard SDAR-algorithm.
Examples
# Synthesize a test record resembling EN AW-6060-T66.
# Explicitly set names to "strain" and "stress".
Al_6060_T66 <- synthesize_test_data(
slope = 69000,
yield.y = 160,
ultimate.y = 215,
ultimate.x = 0.08,
x.name = "strain",
y.name = "stress",
toe.start.y = 3, toe.end.y = 10,
toe.start.slope = 13600
)
# use sdar_lazy() to analyze the (noise-free) synthetic test record
# will print a report and give a plot of the final fit
# \donttest{
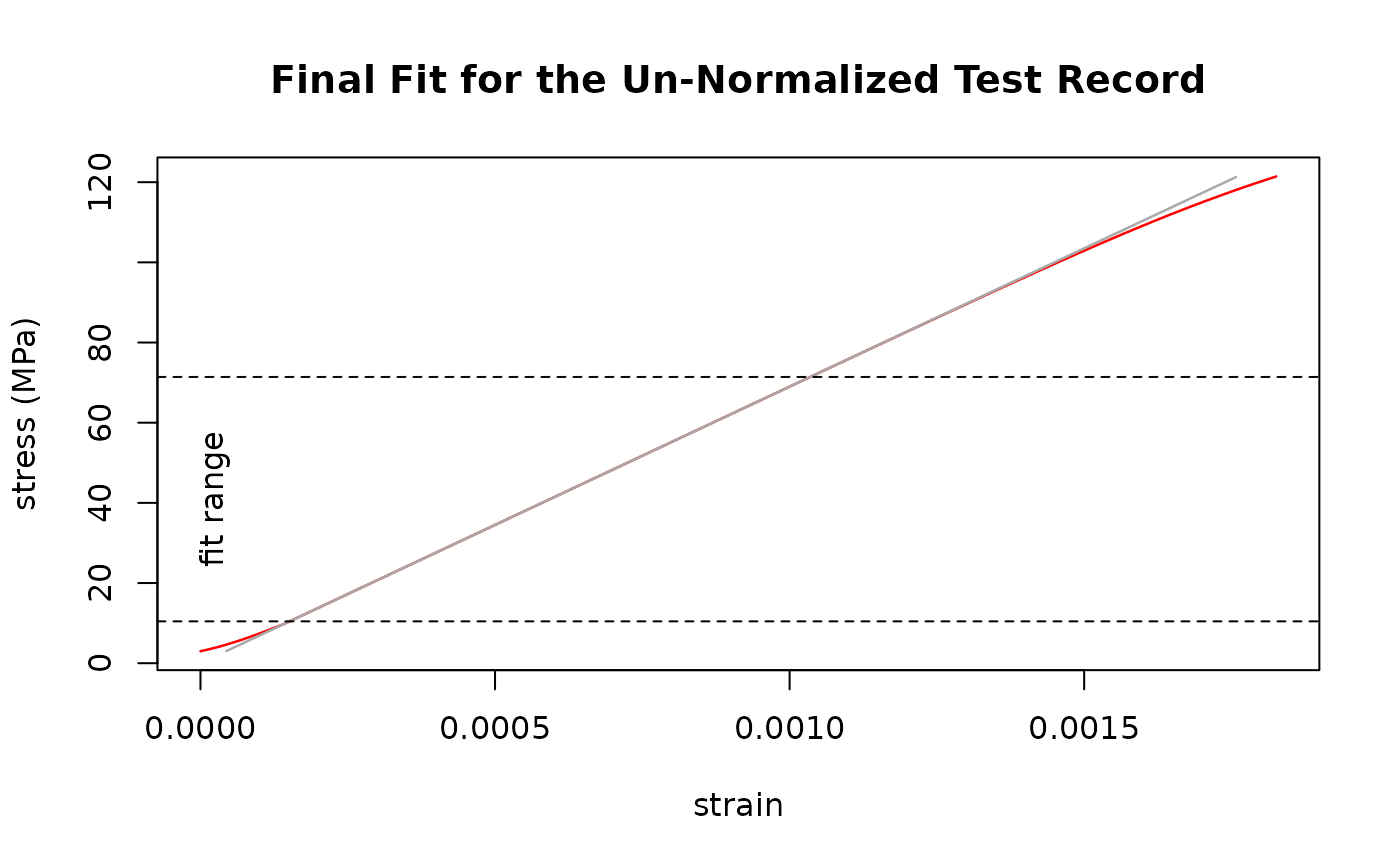
result <- sdar_lazy(Al_6060_T66, strain, stress)
#> Determination of Slope in the Linear Region of a Test Record:
#> Random sub-sampling modification of the SDAR-algorithm
#> Random sub-sampling information:
#> 121 points of 375 points in the normalized range were used.
#> 0 % of the sub-samples passed the data quality checks.
#> 100 % of the sub-samples passed the fit quality checks.
#> 0 % of the sub-samples passed all quality checks.
#>
#> Data Quality Metric: Digital Resolution
#> x
#> Relative x-Resolution: 0.333333333333333
#> % at this resolution: 0
#> % in zeroth bin: 100
#> --> pass
#> y
#> Relative y-Resolution: 0.333333333333333
#> % at this resolution: 0.804289544235925
#> % in zeroth bin: 99.1957104557641
#> --> pass
#> Data Quality Metric: Noise
#> x
#> Relative x-Noise: 1.14246654063749e-14
#> --> pass
#> y
#> Relative y-Noise: 0.0619676935307803
#> --> pass
#> Fit Quality Metric: Curvature
#> 1st Quartile
#> Relative Residual Slope: 0.00086452510296137
#> Number of Points: 46
#> --> pass
#> 4th Quartile
#> Relative Residual Slope: -0.00711062691360424
#> Number of Points: 46
#> --> pass
#> Fit Quality Metric: Fit Range
#> relative fit range: 0.777012483857081
#> --> pass
#> Un-normalized fit
#> Final Slope: 68997.0676230752 MPa
#> True Intercept: 0.00127687293251233 MPa
#> y-Range: 10.445556640625 MPa - 71.4129638671875 MPa
 # }
# }